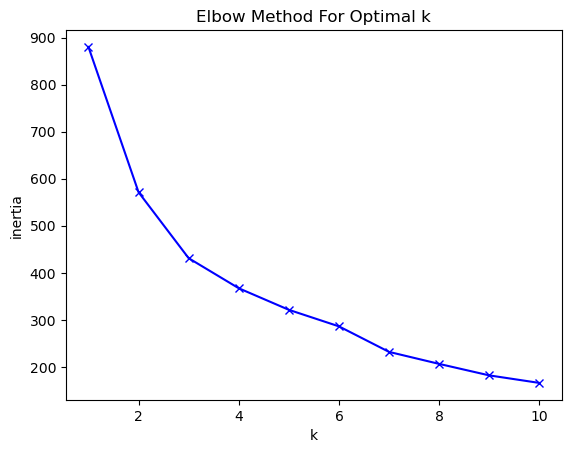
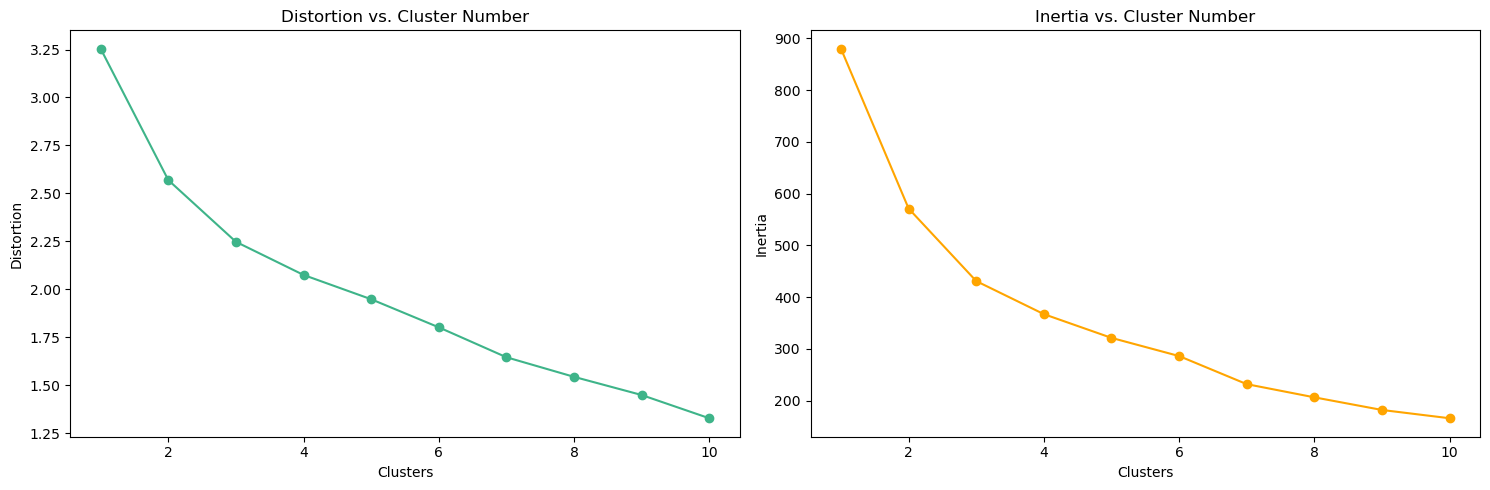
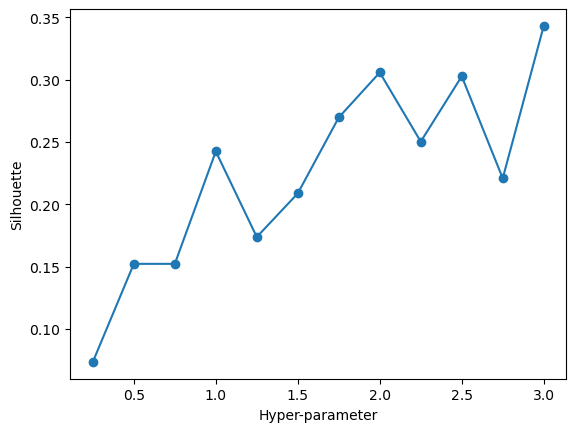
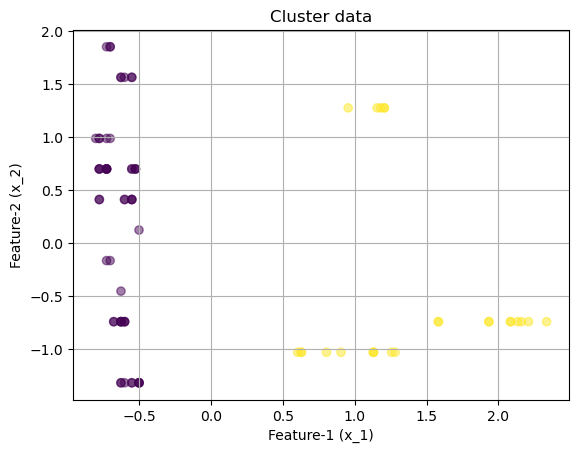
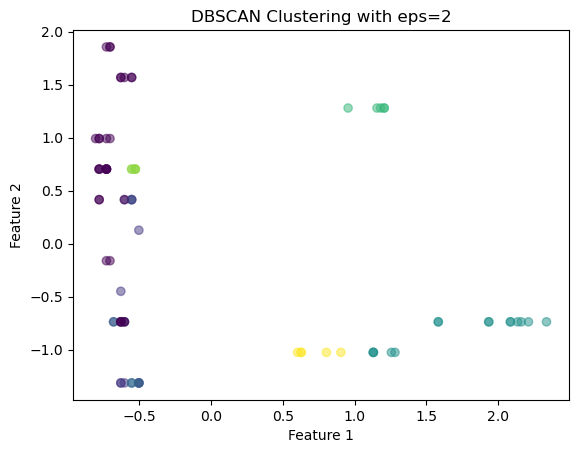
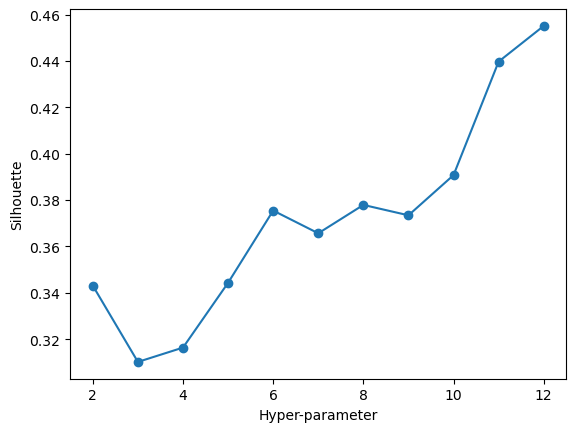
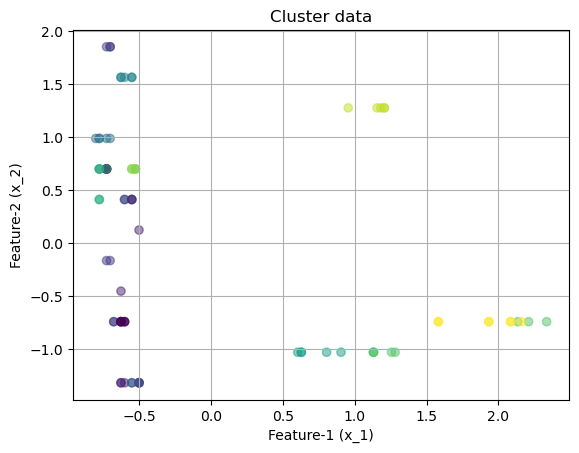
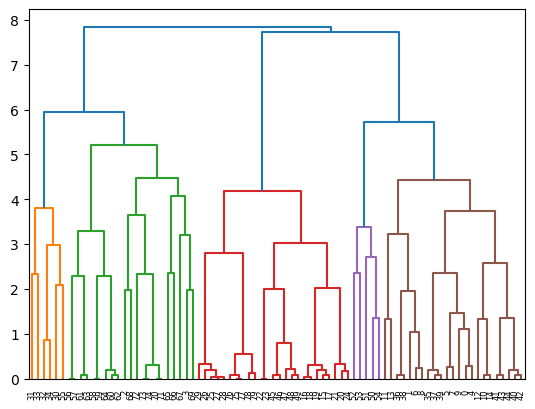
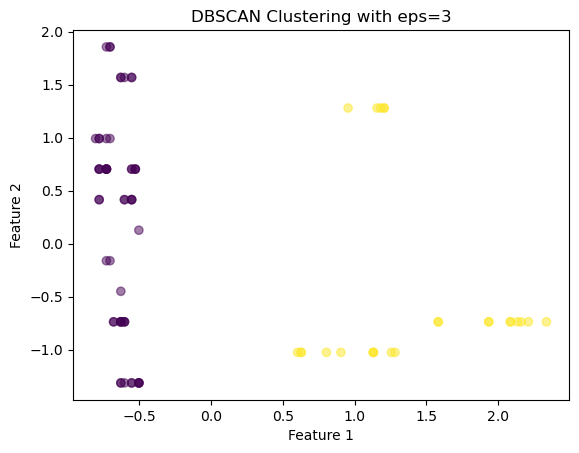
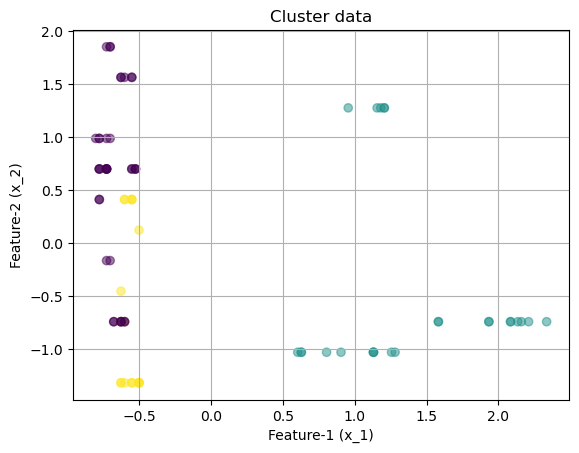
array([[-0.62883646, -0.73831341, -0.61329244, -1.17319439, -0.9119434 ,
0.13303802, -0.5762842 , 1.25937568, -0.97618706, -0.55809982,
-0.90149708],
[-0.60370813, -0.73831341, -0.58645251, -1.17319439, -0.9119434 ,
0.13303802, -0.54706827, 1.25937568, -0.97618706, 1.79179416,
-0.90149708],
[-0.67909312, -0.73831341, -0.66697231, 0.52485012, 0.06434637,
0.13303802, -0.69314792, 1.25937568, -0.97618706, -0.55809982,
-0.90149708],
[-0.67909312, -0.73831341, -0.69381224, 0.52485012, 0.06434637,
0.13303802, -0.69314792, 1.25937568, -0.97618706, 1.79179416,
-0.90149708],
[-0.62883646, -0.4501911 , -0.61329244, -1.17319439, -0.9119434 ,
0.13303802, -0.5762842 , 1.25937568, -0.97618706, -0.55809982,
-0.90149708],
[-0.50319481, -1.31455802, -0.53277265, -1.17319439, -1.64416072,
0.13303802, -0.5762842 , 1.25937568, -0.54232614, -0.55809982,
-0.90149708],
[-0.50319481, -1.31455802, -0.47909279, -1.17319439, -1.64416072,
0.13303802, -0.43020456, 1.25937568, -0.54232614, 1.79179416,
-0.90149708],
[-0.50319481, -1.31455802, -0.50593272, -1.17319439, -1.40008828,
0.13303802, -0.45942048, 1.25937568, -0.54232614, -0.55809982,
-0.90149708],
[-0.50319481, -1.31455802, -0.47909279, -1.17319439, -1.40008828,
0.13303802, -0.45942048, 1.25937568, -0.54232614, 1.79179416,
-0.90149708],
[-0.50319481, 0.12605351, -0.50593272, -1.17319439, -1.40008828,
0.13303802, -0.45942048, 1.25937568, -0.54232614, -0.55809982,
-0.90149708],
[-0.55345147, 0.41417581, -0.55961258, -1.17319439, -1.15601584,
0.13303802, -0.5762842 , 1.25937568, 0.54232614, -0.55809982,
-0.61681379],
[-0.55345147, 0.41417581, -0.53277265, -1.17319439, -1.15601584,
0.13303802, -0.54706827, 1.25937568, 0.54232614, 1.79179416,
-0.61681379],
[-0.60370813, 0.41417581, -0.61329244, -1.17319439, -1.15601584,
-1.19734219, -0.63471606, 1.25937568, 0.54232614, -0.55809982,
-0.61681379],
[-0.60370813, 0.41417581, -0.61329244, -1.17319439, -1.15601584,
-1.19734219, -0.63471606, 1.25937568, 0.54232614, 1.79179416,
-0.61681379],
[-0.55345147, 0.41417581, -0.55961258, -1.17319439, -1.15601584,
0.13303802, -0.5762842 , 1.25937568, 0.54232614, -0.55809982,
-0.52191936],
[ 1.13014664, -1.02643571, 1.21182293, 0. , 0. ,
1.46341823, 1.29353526, 0.74270874, 0.10846523, -0.55809982,
0.90149708],
[ 1.28091662, -1.02643571, 1.37286252, 0. , 0. ,
1.46341823, 1.49804676, 0.74270874, 0.10846523, -0.55809982,
0.99639151],
[ 1.13014664, -1.02643571, 1.21182293, 0. , 0. ,
1.46341823, 1.29353526, 0.74270874, 0.10846523, -0.55809982,
0.99639151],
[ 1.25578829, -1.02643571, 1.34602259, 0. , 0. ,
1.46341823, 1.46883083, 0.74270874, 0.10846523, -0.55809982,
0.99639151],
[ 1.13014664, -1.02643571, 1.21182293, 0. , 0. ,
1.46341823, 1.29353526, 0.74270874, 0.10846523, -0.55809982,
1.09128594],
[ 2.21066483, -0.73831341, 2.20490041, 0. , 0. ,
1.46341823, 2.17001313, 0.74270874, -0.10846523, -0.55809982,
1.37596923],
[ 2.33630648, -0.73831341, 2.31226014, 0. , 0. ,
1.46341823, 2.25766092, 0.74270874, -0.10846523, -0.55809982,
1.47086366],
[ 1.9342532 , -0.73831341, 1.93650109, 0. , 0. ,
-1.19734219, 1.9362857 , 0.74270874, -0.10846523, -0.55809982,
1.47086366],
[ 1.9342532 , -0.73831341, 1.93650109, 0. , 0. ,
-1.19734219, 1.9362857 , 0.74270874, -0.10846523, -0.55809982,
1.47086366],
[ 2.13527984, -0.73831341, 2.12438061, 0. , 0. ,
1.46341823, 2.08236535, 0.74270874, -0.10846523, -0.55809982,
1.47086366],
[ 1.1804033 , 1.27854273, 1.29234272, 0. , 0. ,
-1.19734219, 1.41039898, 0.74270874, 0.32539569, -0.55809982,
1.2810748 ],
[ 1.15527497, 1.27854273, 1.21182293, 0. , 0. ,
-1.19734219, 1.29353526, 0.74270874, 0.32539569, -0.55809982,
1.2810748 ],
[ 1.20553163, 1.27854273, 1.31918265, 0. , 0. ,
-1.19734219, 1.43961491, 0.74270874, 0.32539569, -0.55809982,
1.2810748 ],
[ 1.20553163, 1.27854273, 1.31918265, 0. , 0. ,
-1.19734219, 1.41039898, 0.74270874, 0.32539569, -0.55809982,
1.2810748 ],
[ 0.95424833, 1.27854273, 1.13130313, 0. , 0. ,
-1.19734219, 1.35196712, 0.74270874, 0.32539569, -0.55809982,
1.2810748 ],
[-0.72934978, 0.99042042, -0.77433204, 0.52485012, 1.16267236,
1.46341823, -0.83922756, -1.32395905, 0.7592566 , -0.55809982,
-0.80660265],
[-0.70422145, 0.99042042, -0.72065217, 0.52485012, 1.16267236,
1.46341823, -0.7807957 , -1.32395905, 0.7592566 , 1.79179416,
-0.80660265],
[-0.77960644, 0.99042042, -0.8280119 , 2.22289464, 2.74914323,
1.46341823, -0.86844349, -1.32395905, 0.7592566 , -0.55809982,
-0.80660265],
[-0.77960644, 0.99042042, -0.80117197, 2.22289464, 2.74914323,
1.46341823, -0.83922756, -1.32395905, 0.7592566 , 1.79179416,
-0.80660265],
[-0.80473477, 0.99042042, -0.85485183, 2.22289464, 3.60339678,
1.46341823, -0.89765942, -1.32395905, 0.7592566 , -0.55809982,
-0.80660265],
[-0.60370813, -1.31455802, -0.61329244, -1.17319439, -0.66787096,
0.13303802, -0.5762842 , -0.29062516, -1.19311752, -0.55809982,
-0.80660265],
[-0.55345147, -1.31455802, -0.53277265, -1.17319439, -0.66787096,
0.13303802, -0.51785234, -0.29062516, -1.19311752, 1.79179416,
-0.80660265],
[-0.62883646, -1.31455802, -0.61329244, -1.17319439, -0.66787096,
0.13303802, -0.5762842 , -0.29062516, -1.19311752, -0.55809982,
-0.71170822],
[-0.55345147, -1.31455802, -0.53277265, -1.17319439, -0.66787096,
0.13303802, -0.51785234, -0.29062516, -1.19311752, 1.79179416,
-0.71170822],
[-0.62883646, -1.31455802, -0.61329244, -1.17319439, -0.66787096,
0.13303802, -0.5762842 , -0.29062516, -1.19311752, -0.55809982,
-0.61681379],
[-0.52832314, 0.70229812, -0.53277265, -1.17319439, -0.54583473,
0.13303802, -0.54706827, -0.29062516, 0.97618706, -0.55809982,
0.52191936],
[-0.55345147, 0.70229812, -0.55961258, -1.17319439, -0.54583473,
-1.19734219, -0.5762842 , -0.29062516, 0.97618706, -0.55809982,
0.52191936],
[-0.52832314, 0.70229812, -0.53277265, -1.17319439, -0.54583473,
0.13303802, -0.54706827, -0.29062516, 0.97618706, -0.55809982,
0.61681379],
[-0.55345147, 0.70229812, -0.55961258, -1.17319439, -0.54583473,
-1.19734219, -0.5762842 , -0.29062516, 0.97618706, -0.55809982,
0.61681379],
[-0.52832314, 0.70229812, -0.53277265, -1.17319439, -0.54583473,
0.13303802, -0.54706827, -0.29062516, 0.97618706, -0.55809982,
0.71170822],
[ 1.58245658, -0.73831341, 1.48022225, 0. , 0. ,
0.13303802, 1.35196712, -0.29062516, -0.32539569, -0.55809982,
0.80660265],
[ 1.58245658, -0.73831341, 1.48022225, 0. , 0. ,
0.13303802, 1.35196712, -0.29062516, -0.32539569, -0.55809982,
0.90149708],
[ 2.16040817, -0.73831341, 1.90966116, 0. , 0. ,
0.13303802, 1.64412641, -0.29062516, -0.32539569, -0.55809982,
0.99639151],
[ 2.08502318, -0.73831341, 1.88282123, 0. , 0. ,
0.13303802, 1.61491048, -0.29062516, -0.32539569, -0.55809982,
1.09128594],
[ 2.08502318, -0.73831341, 1.88282123, 0. , 0. ,
0.13303802, 1.61491048, -0.29062516, -0.32539569, -0.55809982,
1.18618037],
[-0.62883646, 1.56666503, -0.64013238, 0.52485012, -0.30176229,
1.46341823, -0.60550013, -1.840626 , -1.62697843, -0.55809982,
-1.56575808],
[-0.60370813, 1.56666503, -0.61329244, 0.52485012, -0.30176229,
1.46341823, -0.5762842 , -1.840626 , -1.62697843, 1.79179416,
-1.56575808],
[-0.55345147, 1.56666503, -0.55961258, -1.17319439, -1.40008828,
0.13303802, -0.63471606, -1.840626 , -1.62697843, -0.55809982,
-1.75554694],
[-0.55345147, 1.56666503, -0.53277265, -1.17319439, -1.40008828,
0.13303802, -0.54706827, -1.840626 , -1.62697843, 1.79179416,
-1.75554694],
[-0.62883646, 1.56666503, -0.64013238, 0.52485012, -0.30176229,
0.13303802, -0.60550013, -1.840626 , -1.62697843, -0.55809982,
-1.75554694],
[-0.77960644, 0.70229812, -0.80117197, 2.22289464, 1.77285347,
1.46341823, -0.86844349, -1.840626 , 1.41004798, -0.55809982,
-0.23723607],
[-0.72934978, 0.70229812, -0.7474921 , 0.52485012, 0.06434637,
-1.19734219, -0.7807957 , -1.840626 , 1.41004798, 1.79179416,
-0.14234164],
[-0.72934978, 0.70229812, -0.7474921 , 0.52485012, 0.06434637,
-1.19734219, -0.7807957 , -1.840626 , 1.41004798, 1.79179416,
-0.14234164],
[-0.72934978, 0.70229812, -0.7474921 , 0.52485012, 0.06434637,
-1.19734219, -0.7807957 , -1.840626 , 1.41004798, -0.55809982,
-0.14234164],
[-0.72934978, 0.70229812, -0.7474921 , 0.52485012, 0.06434637,
-1.19734219, -0.7807957 , -1.840626 , 1.41004798, -0.55809982,
-0.14234164],
[-0.62883646, -0.73831341, -0.61329244, 0.52485012, 0.06434637,
0.13303802, -0.5762842 , -0.8072921 , 1.62697843, -0.55809982,
-0.61681379],
[-0.60370813, -0.73831341, -0.61329244, 0.52485012, 0.06434637,
0.13303802, -0.63471606, -0.8072921 , 1.62697843, 1.79179416,
-0.61681379],
[-0.62883646, -0.73831341, -0.61329244, 0.52485012, 0.06434637,
0.13303802, -0.5762842 , -0.8072921 , 1.62697843, -0.55809982,
-0.52191936],
[-0.60370813, -0.73831341, -0.61329244, 0.52485012, 0.06434637,
0.13303802, -0.63471606, -0.8072921 , 1.62697843, 1.79179416,
-0.52191936],
[-0.62883646, -0.73831341, -0.61329244, 0.52485012, 0.06434637,
0.13303802, -0.60550013, -0.8072921 , 1.62697843, -0.55809982,
-0.42702493],
[-0.70422145, 1.85478734, -0.72065217, 0.52485012, 0.7965637 ,
1.46341823, -0.69314792, 0.22604179, -1.41004798, -0.55809982,
-0.80660265],
[-0.70422145, 1.85478734, -0.69381224, 0.52485012, 0.7965637 ,
1.46341823, -0.66393199, 0.22604179, -1.41004798, 1.79179416,
-0.80660265],
[-0.72934978, 1.85478734, -0.7474921 , 0.52485012, 0.7965637 ,
-1.19734219, -0.72236385, 0.22604179, -1.41004798, 1.79179416,
-0.80660265],
[-0.70422145, -0.1620688 , -0.72065217, 0.52485012, 0.7965637 ,
1.46341823, -0.69314792, 0.22604179, -1.41004798, -0.55809982,
-0.80660265],
[-0.72934978, -0.1620688 , -0.7474921 , 0.52485012, 0.7965637 ,
-1.19734219, -0.72236385, 0.22604179, -1.41004798, 1.79179416,
-0.80660265],
[-0.77960644, 0.41417581, -0.80117197, 2.22289464, 1.89488969,
-1.19734219, -0.83922756, 0.22604179, -0.7592566 , -0.55809982,
0.04744721],
[-0.77960644, 0.41417581, -0.80117197, 2.22289464, 1.89488969,
-1.19734219, -0.83922756, 0.22604179, -0.7592566 , -0.55809982,
0.04744721],
[-0.72934978, 0.70229812, -0.77433204, 0.52485012, 0.30841881,
-1.19734219, -0.81001163, 0.22604179, -0.7592566 , -0.55809982,
0.14234164],
[-0.77960644, 0.70229812, -0.80117197, 2.22289464, 1.89488969,
-1.19734219, -0.83922756, 0.22604179, -0.7592566 , -0.55809982,
0.14234164],
[-0.77960644, 0.70229812, -0.80117197, 2.22289464, 1.89488969,
-1.19734219, -0.83922756, 0.22604179, -0.7592566 , -0.55809982,
0.14234164],
[ 0.62758004, -1.02643571, 0.67502429, 0. , 0. ,
-1.19734219, 0.73843261, 0.22604179, 1.19311752, -0.55809982,
1.66065251],
[ 0.60245171, -1.02643571, 0.64818436, 0. , 0. ,
-1.19734219, 0.65078482, 0.22604179, 1.19311752, -0.55809982,
1.66065251],
[ 0.62758004, -1.02643571, 0.67502429, 0. , 0. ,
-1.19734219, 0.73843261, 0.22604179, 1.19311752, -0.55809982,
1.66065251],
[ 0.90399167, -1.02643571, 0.94342361, 0. , 0. ,
-1.19734219, 1.00137597, 0.22604179, 1.19311752, -0.55809982,
1.75554694],
[ 0.80347835, -1.02643571, 0.88974374, 0. , 0. ,
-1.19734219, 1.0305919 , 0.22604179, 1.19311752, -0.55809982,
1.75554694]])